Strain Tracking and Engraftment
Understanding the dynamics of specific microbial strains is crucial for many microbiome studies, especially when tracking how these strains change over time or across different environments. Our Clonal-Level Microbiomics™ offers the most sensitive and accurate microbiome analysis available, providing detailed information about clonal populations (i.e., strains) in your samples based on single nucleotide variations (SNVs).
Strain Tracking and Engraftment
Understanding the dynamics of specific microbial strains is crucial for many microbiome studies, especially when tracking how these strains change over time or across different environments. Our Clonal-Level Microbiomics™ offers the most sensitive and accurate microbiome analysis available, providing detailed information about clonal populations (i.e., strains) in your samples based on single nucleotide variations (SNVs).

Why Strain-Level Analysis Matters
Standard microbiome analysis methods often lack the resolution to differentiate between closely related strains within the same species. However, strain-level differences can have significant implications in various research areas:
-
Fecal Microbiota Transplantation (FMT): Determine which strains in the recipient originated from the donor, even when both have endogenous microbes of the same species.
-
Live Biotherapeutic Products (LBPs) and Probiotics: Track the engraftment and persistence of administered strains in the host microbiome.
-
Clinical Research: Link specific microbial strains to health outcomes or disease states.
-
Environmental Studies: Monitor strain dynamics across different environments or over time.
Our Advanced Methodology
Our robust technique leverages variations at the single nucleotide level to achieve clonal-level resolution. Here's how our method stands out:
High Precision
Resolve strains with an average nucleotide identity (ANI) of 99.9%, allowing differentiation between nearly identical strains.
Strain Tracking
Identify and monitor specific strains across samples collected before and after treatment or intervention.
Phylogenetic Analysis
Build phylogenetic trees to organize strains according to their genetic relatedness.
Functional Insights
Link groups of related strains (clades) to clinical information and identify genes responsible for functional differences.
Mechanistic Understanding
Support correlations with clearer hypotheses about modes of action affecting host health outcomes.
Applications of Clonal-Level Profiling
Our Clonal-Level Microbiome Profiling™ is particularly powerful for:
-
Engraftment Analysis: Track donor strains from FMT, LBPs, or probiotics in post-treatment samples to assess engraftment success.
-
Therapeutic Development: Evaluate how specific strains contribute to treatment efficacy or patient response.
-
Personalized Medicine: Identify individual-specific strains that may influence health or disease progression.
-
Microbial Ecology: Study strain-level dynamics in microbial communities within various environments.

Clonal-Level Microbiomics™ for strain tracking
In some studies, researchers may want to detect the presence of microbial strains in samples over time. This tracking of strains is important, for example, in determining whether a probiotic strain persists in the gut or whether strains from a fecal microbiota transplant have stably engrafted in their new environment. Many analysis pipelines, however, do not provide sufficient resolution to detect strains (or bacterial clones), which reduces confidence in determining whether the exact strains of interest are present over time.
Cmbio offers its Clonal-level Microbiome Profiling pipeline, providing the most sensitive and accurate microbiome profiling in the field. With Clonal-level Microbiomics™️, we resolve clonal populations (i.e. strains) found in samples using single nucleotide differences, and use phylogeny to relate them to each other and build trees. The pipeline can resolve strains with an average nucleotide identity (ANI) of 99.9%. This method also enables us to link host or microbial traits to clades (phylogenetic groups) within microbial populations, and even to mine the genes behind the functional differences.
Tracking Bacterial Strains from Mother to Infant
Fecal microbiota transplantation (FMT) from a donor is an effective treatment for individuals with recurrent Clostridioides difficile infection (CDI). The donor FMT population often engrafts in the recipient, but only with high-resolution microbiome analysis is it possible to determine whether the exact strains from the donor are present in the recipient or in another environment. If researchers observe highly similar strains (or clones) in multiple environments, the likelihood is high that transfer has occurred from one environment to another.
The Cmbio team collaborated with researchers from the Technical University of Denmark and the Aarhus University Hospital on a project that aimed to determine whether strains of bacteria introduced to a pregnant individual through an FMT intervention were transferred to her healthy infant born 26 weeks later.
Study Design & Analysis
The case report describes a detailed analysis that aimed to discover whether bacterial strain transfer occurred from the FMT donor to the pregnant mother, and further, to her infant born at term. The mother developed CDI during pregnancy and was treated successfully with a single dose of FMT before giving vaginal birth at term to a healthy infant. For the analysis, fecal samples were collected from all three individuals: the FMT donor; the mother at several time points (before FMT, and 1, 8, 15, 22, 26, and 50 weeks after FMT); and the infant at several time points (meconium at birth and 3 & 6 months after birth).
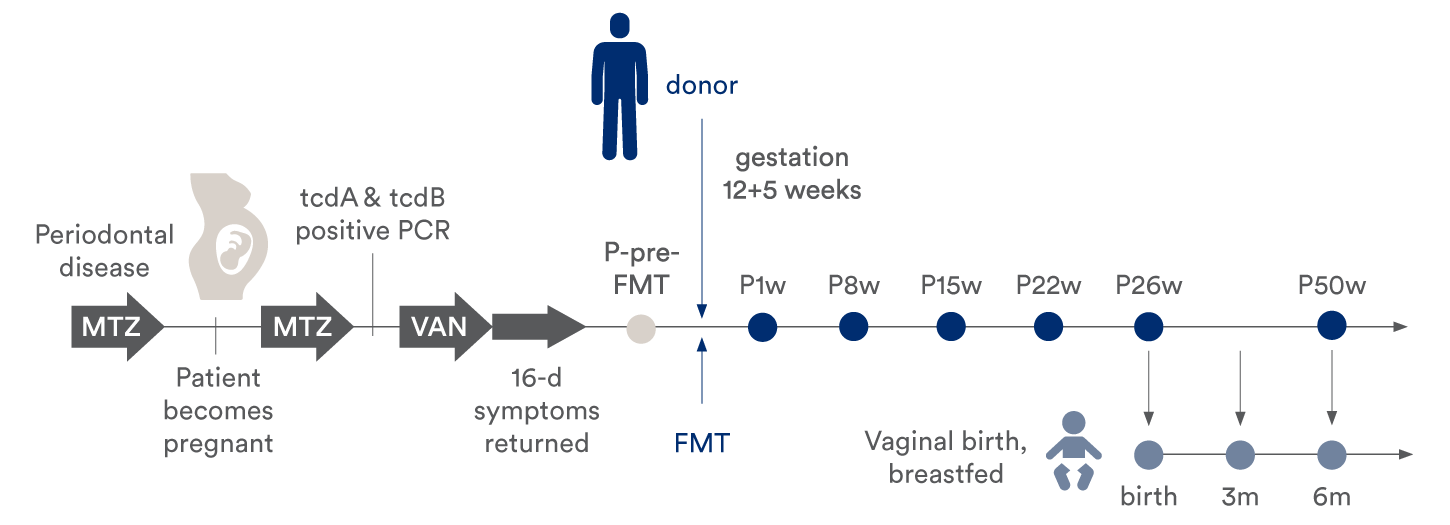 Timeline indicating treatment, FMT, sampling time points for the mother and infant. Sampling time was relative to FMT (for the patient) or time of birth (for the infant). The patient vaginally delivered a healthy child at term, 26 weeks after the FMT.
Timeline indicating treatment, FMT, sampling time points for the mother and infant. Sampling time was relative to FMT (for the patient) or time of birth (for the infant). The patient vaginally delivered a healthy child at term, 26 weeks after the FMT.
Samples were analyzed using Cmbio’s Clonal-level Microbiomics™️ platform. Single nucleotide variants were identified in the metagenomic samples and were compared to a collection of samples from healthy infants and adults. Two forms of engraftment profiling were used, allowing researchers to track microbial populations from FMT donor to recipient—despite the recipient already having closely-related endogenous strains.
Study Results
Results showed that, before receiving FMT, the pregnant mother’s gut microbiota was characterized by low diversity and the presence of potential pathogens. Upon receiving FMT, these features diminished. After the infant’s birth, the precise bacterial strains administered to the mother via FMT were found in the infant, suggesting both persistence over time and cross-generational transfer of the strains.
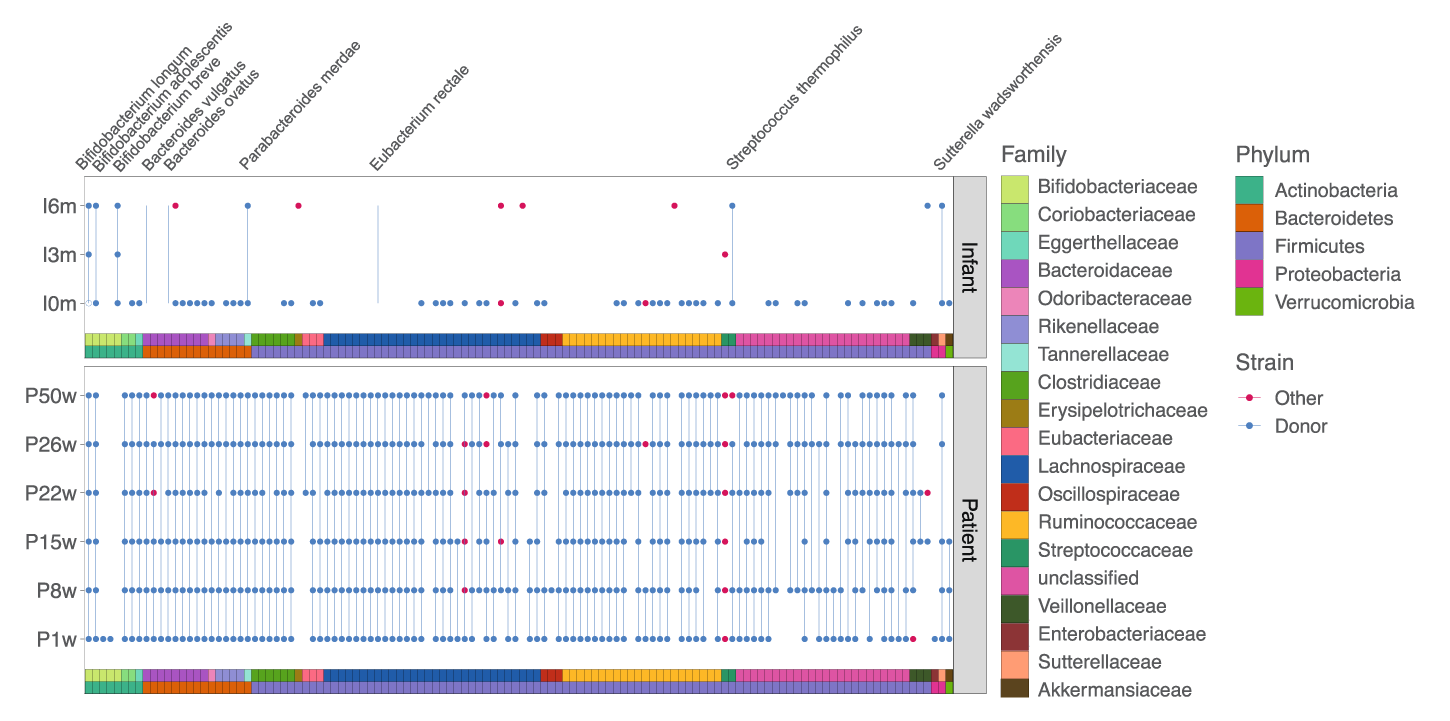
Detection of donor (blue dots) and non-donor (red dots) strains longitudinally in the mother and infant. None of these strains were detected in the mother pre-FMT.
See the study published in *Microbiome.* ¹
This case study demonstrates the potential to track precise strains from one environment to another with confidence, and to determine the persistence of these strains over time. Vertical transfer of bacterial strains from a pregnant mother to an infant was observed, demonstrating a potential route for influencing the infant gut microbiota.
Let’s find the answers. Let our expert scientists guide you. Contact us to discuss your study design and research needs on engraftment and strain tracking.
Reference
¹ Wei, S., Jespersen, M.L., Baunwall, S.M.D. et al. Cross-generational bacterial strain transfer to an infant after fecal microbiota transplantation to a pregnant patient: a case report. Microbiome 10, 193 (2022). https://doi.org/10.1186/s40168-022-01394-w