Multi-omics Data Integration and Analysis
No single analysis can account for how the microbiome influences the host in health or disease. Multi-omics integrative analysis brings together different types of data to help understand the system as a whole and how the microbiome works in this context to influence the host. In the hands of a highly experienced team, such analyses can avoid spurious correlations and identify true signals that can be used for drug development or other important applications.
The Clinical Microbiomics Clonal-level Microbiome Profiling pipeline provides the most sensitive and accurate microbiome profiling in the field, with clonal-level resolution. We offer development and curation of prior knowledge ontologies, enabling smart dimensionality reduction. Gene-neighborhood-aware functional annotation is part of the Clonal-level Microbiomics™️ Platform to identify and/or infer functionality of all species in a community.
Case Study: Targets for Insulin Resistance
Insulin resistance precedes more serious metabolic problems such as ischemic cardiovascular disease and type 2 diabetes. A growing body of evidence shows gut microbiota differences, as well as an altered metabolite pool, in those with insulin resistance. The current challenge in the field is to identify the mechanisms involved and pinpoint key bacteria and metabolites responsible for the insulin resistance phenotype.
Our team collaborated with researchers at the University of Copenhagen and Novo Nordisk Foundation on a study that aimed to identify human drug development targets with the potential to diminish insulin resistance and incidence of associated metabolic and cardiovascular disorders.
Study design & analysis
In this study, we collaborated to integrate shotgun sequencing gut microbiome data, untargeted serum metabolome data, and measures of host physiology for 277 non-diabetic individuals. This yielded a dataset with 325 polar metabolites, 876 molecular lipids, and 7 million microbial genes. We first reduced dimensionality of the data, then filtered for phenotype-relevant features. Subsequently, we carried out cross-domain correlation/association analyses.
Study results
Our results showed insulin resistance in these subjects was driven primarily by the bacterium P. copri, with elevated branched-chain amino acids and/or breakdown byproducts aggravating glucose intolerance and leading to insulin resistance. The insulin resistance and glucose intolerance phenotype was recapitulated in mice when fed a high-fat diet and challenged with P. copri, providing evidence for causation.
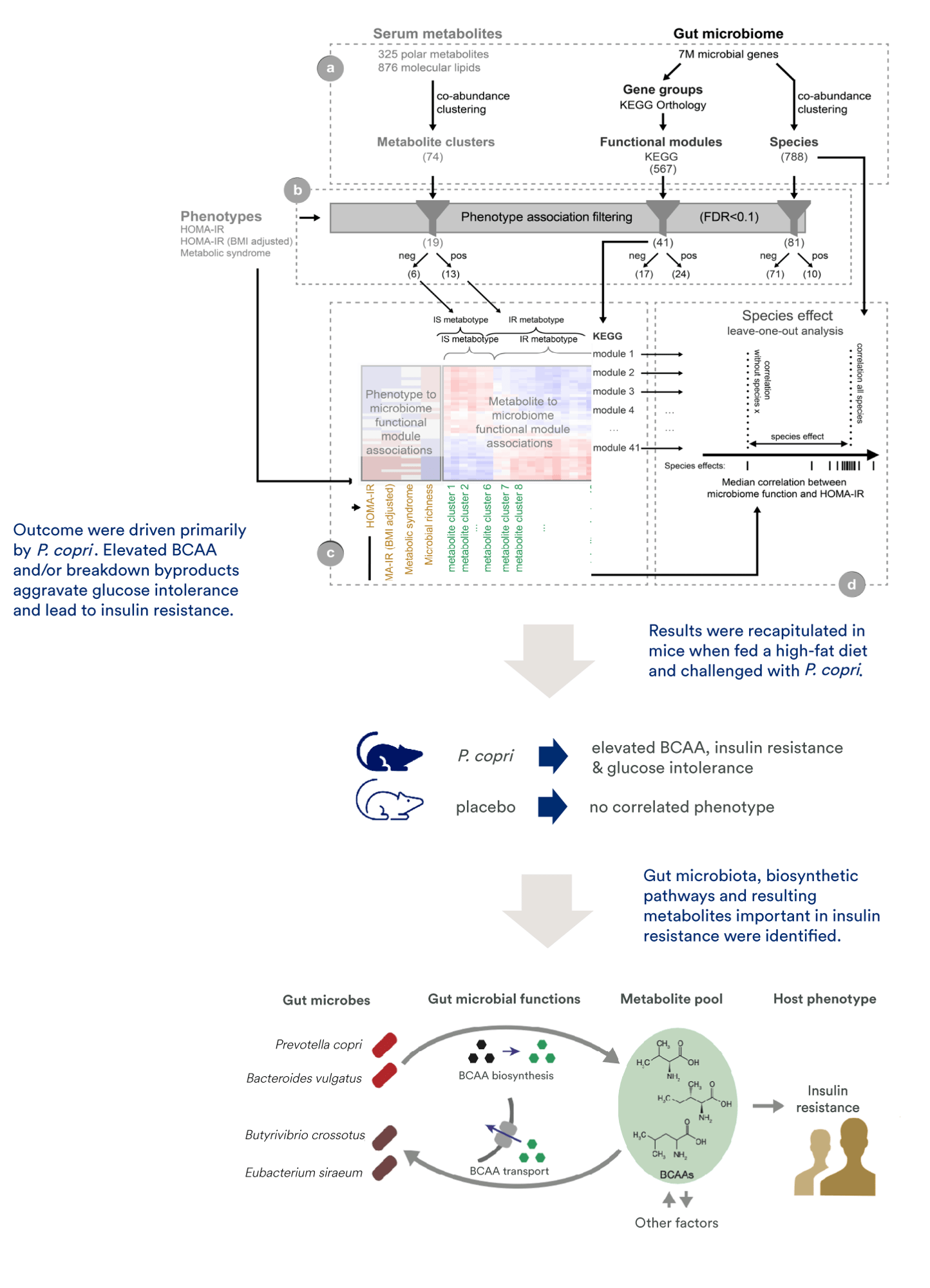
The study and follow-up work provided robust evidence that insulin resistance may be induced by gut microbiome-derived metabolites. A central advantage of this approach is the successful integration of prior knowledge to overcome functional redundancy across microbiome species.
See the study published in Nature. ¹
See the computational framework published in Nature Protocols. ²
This case study demonstrates the possibility of connecting microbiome and other omics readouts to aspects of host physiology, to successfully identify potential mechanistic links that can be tested further in pre-clinical and clinical settings. Combining datasets enables a comprehensive understanding of bacteria to facilitate strain selection and de-risking prior to assay-based characterizations or regulatory submissions.
Let’s find the answers. Let our expert scientists guide you. Contact us to discuss your study design and research needs for integrative systems biology.
References
¹ Pedersen, H., Gudmundsdottir, V., Nielsen, H. et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016). https://doi.org/10.1038/nature18646
² Pedersen, H.K., Forslund, S.K., Gudmundsdottir, V. et al. A computational framework to integrate high-throughput ‘-omics’ datasets for the identification of potential mechanistic links. Nature Protocols 13, 2781–2800 (2018). https://doi.org/10.1038/s41596-018-0064-z