StrainQTM
Our strain tracking and engraftment pipeline, StrainQTM, offers the most sensitive and accurate clonal-level microbiome analysis available.
Strain Tracking and Engraftment
Understanding the dynamics of specific microbial strains is crucial for many microbiome studies, especially when tracking how these strains change over time or across different environments. Our Clonal-Level Microbiomics™ offers the most sensitive and accurate microbiome analysis available, providing detailed information about clonal populations (i.e., strains) in your samples based on single nucleotide variations (SNVs).
Ready to advance your microbiome research?
Contact our scientists today!
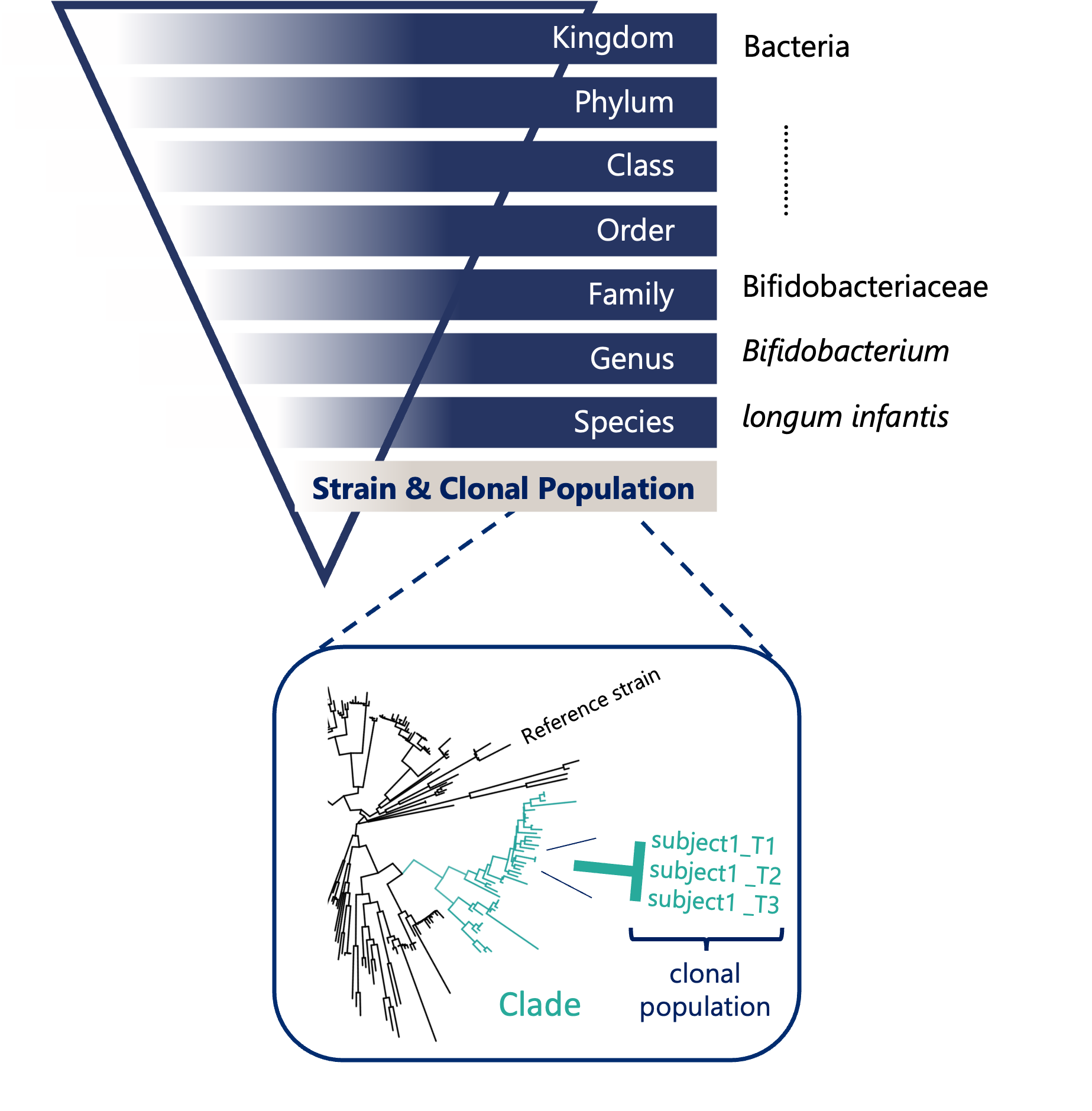
What is clonal-level profiling?
When we talk about the microbiome, it’s easy to imagine a collection of bacteria, each neatly categorized by species. However, the reality is far more nuanced: every individual harbors a unique collection of microbial strains. Within a single microbial species, there can be countless strains-each with subtle genetic differences that can have profound effects on their function. These differences influence everything from how microbes interact with your immune system to how they metabolize nutrients or resist antibiotics.
This is where clonal-level profiling comes in: simply comparing a sample to a reference database overlooks important diversity that is unique to your sample. Instead, our denovo approach can distinguish between these closely related strains and understand their unique roles within the microbiome.
Why StrainQTM is different?
Most traditional methods, such as 16S rRNA sequencing, are limited to species-level resolution and cannot distinguish between closely related strains. Even qPCR primers designed for strains often lacks specificity and detects other strains of the same species. In contrast, Cmbio's robust technique leverages variations at the single nucleotide level to achieve clonal-level resolution. Here's how our method stands out:
High Precision
Resolve strains with an average nucleotide identity (ANI) of 99.9%, allowing differentiation between nearly identical strains.
Superior Sensitivity
88% higher sensitivity than StrainPhlAn in benchmarking, allowing you to detect more engraftment - even of rare and low-abundant strains.
Phylogenetic Analysis
Build phylogenetic trees to organize strains according to their genetic relatedness.
Functional Insights
Link groups of related strains (clades) to clinical information and identify genes responsible for functional differences.
Multi-Strain Populations
Support detection and abundance quantification even when multiple strains of the same species are present.
Engraftment analysis
Clonal-level microbial profiling enables precise tracking of therapeutic strains in the microbiome, revealing true engraftment and guiding the development of targeted, personalized interventions.
What is engraftment analysis?
Understanding which microbial strains successfully engraft - colonize and persist - in a host’s microbiome is critical for evaluating therapies like probiotics, fecal microbiota transplantation (FMT), and live biotherapeutics. Traditional methods, which group microbes at the species level, often overestimate engraftment by conflating administered strains with native ones. Clonal-level profiling solves this by detecting genetic differences down to single nucleotides, ensuring precise tracking. For example, in FMT, it distinguishes donor-derived strains from a recipient’s existing microbes, revealing true engraftment rates that species-level tools miss. Similarly, probiotics can be confirmed to colonize only when their unique genetic signatures persist amid similar native strains.
How does it add value to my study?
This precision links specific strains to functional outcomes, such as bile acid shifts that suppress pathogens like C. difficile or antimicrobial production that modulates gut immunity. Engraftment data also guides intervention design: if a strain fails to persist, dosing can be adjusted; if native microbes block colonization, formulations can include prebiotics to shift the environment. Beyond tracking, clonal-level analysis uncovers mechanisms - whether a therapy works by introducing new strains or amplifying beneficial native ones - accelerating the development of targeted, personalized solutions. In an era of microbiome-based medicine, this resolution transforms guesswork into actionable insight, ensuring therapies are as unique as the microbial communities they aim to restore.
Case Studies
Discover how StrainQTM has been applied in real-world research by reading our case studies, based on publications that demonstrate the effectiveness of our pipeline in strain tracking and engraftment analysis.
Engraftment Analysis of Probiotic in Infants
This peer-reviewed and published study employed our strain-level metagenomic analysis to track the precise colonization and persistence of Bifidobacterium longum subspecies infantis LMG11588 in infant microbiomes.
Strain Differentiation and Colonization Patterns
Using single nucleotide variant (SNV) analysis, we distinguished the supplemented LMG11588 strain from naturally occurring (endogenous) B. infantis strains with remarkable precision. The phylogenetic analysis positioned most supplemented samples (68.2%, n=277) in a distinct "LMG11588 clade" with negligible genetic variability, while remaining samples fell into an "Other B. infantis" clade with higher variability.

Key findings about strain colonization include:
- At baseline, B. infantis was present in 16% of infants across all groups, but none of these were the LMG11588 strain
- During intervention, LMG11588 successfully colonized 78.7% of infants in both experimental groups
- The probiotic strain persisted in 70.3% of these infants even 4 weeks after supplementation ended
- Notably, colonization with LMG11588 did not disrupt native B. infantis strains - endogenous strain colonization patterns remained similar between all groups
Ecological Impact
The study reveals that LMG11588 colonization occurred without ecological disruption:
- Infants colonized with either LMG11588 or "Other B. infantis" showed similar abundances of overall Bifidobacteria and infant-type Bifidobacteria
- Microbial community development trajectories were comparable between infants colonized by LMG11588 and those with autochthonous strains
- This contrasted with infants lacking any B. infantis strains, who showed significantly different community development

The accompanying figures demonstrate the phylogenetic differentiation of LMG11588 from other B. infantis strains, and the distribution patterns of these strains across treatment groups over time, confirming LMG11588's successful engraftment without displacing native strains.
See the published study in Frontiers in Nutrition here.
This study demonstrates the potential to track probiotic strains with confidence, and the insight that it can give. In addition to applying our clonal-level profiling pipeline, we deployed fecal community type analysis and other advances biostatistics. To learn more about how we can support your infant nutrition research read more here.
Reference
Capeding MRZ, Phee LCM, Ming C, et al. Safety, efficacy, and impact on gut microbial ecology of a Bifidobacterium longum subspecies infantis LMG11588 supplementation in healthy term infants: a randomized, double-blind, controlled trial in the Philippines. Front Nutr 10, 1319873 (2023). https://doi.org/10.3389/fnut.2023.1319873
Tracking Bacterial Strains from Mother to Infant
Fecal microbiota transplantation (FMT) from a donor is an effective treatment for individuals with recurrent Clostridioides difficile infection (CDI). The donor FMT population often engrafts in the recipient, but only with high-resolution microbiome analysis is it possible to determine whether the exact strains from the donor are present in the recipient or in another environment. If researchers observe highly similar strains (or clones) in multiple environments, the likelihood is high that transfer has occurred from one environment to another.
The Cmbio team collaborated with researchers from the Technical University of Denmark and the Aarhus University Hospital on a project that aimed to determine whether strains of bacteria introduced to a pregnant individual through an FMT intervention were transferred to her healthy infant born 26 weeks later.
Study Design & Analysis
The case report describes a detailed analysis that aimed to discover whether bacterial strain transfer occurred from the FMT donor to the pregnant mother, and further, to her infant born at term. The mother developed CDI during pregnancy and was treated successfully with a single dose of FMT before giving vaginal birth at term to a healthy infant. For the analysis, fecal samples were collected from all three individuals: the FMT donor; the mother at several time points (before FMT, and 1, 8, 15, 22, 26, and 50 weeks after FMT); and the infant at several time points (meconium at birth and 3 & 6 months after birth).
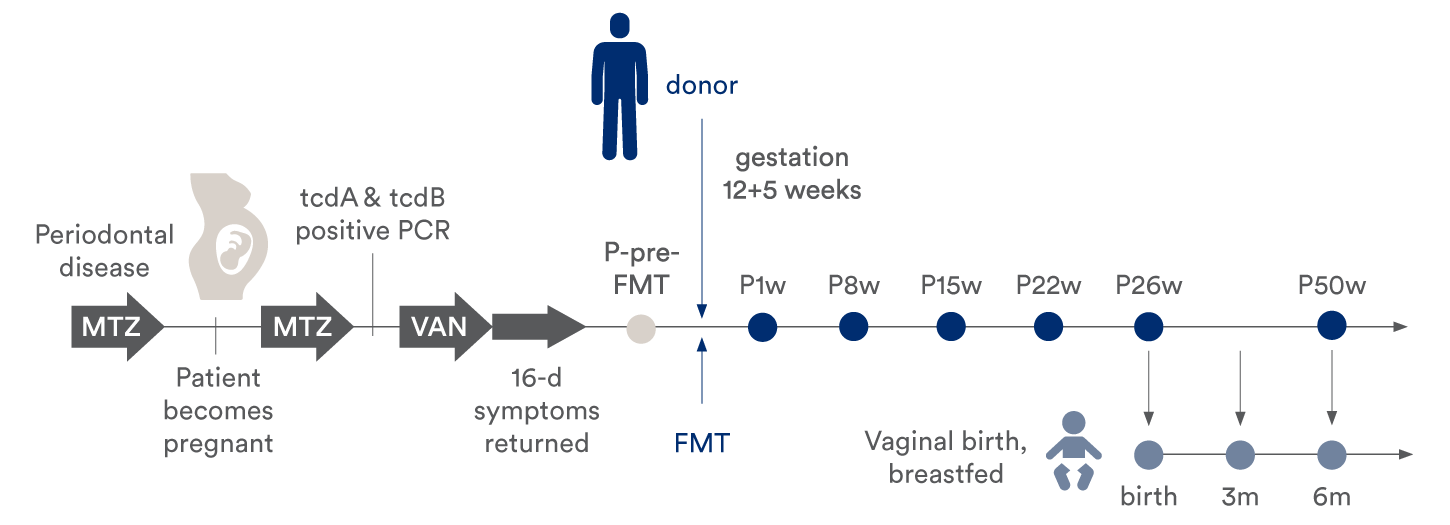 Timeline indicating treatment, FMT, sampling time points for the mother and infant. Sampling time was relative to FMT (for the patient) or time of birth (for the infant). The patient vaginally delivered a healthy child at term, 26 weeks after the FMT.
Timeline indicating treatment, FMT, sampling time points for the mother and infant. Sampling time was relative to FMT (for the patient) or time of birth (for the infant). The patient vaginally delivered a healthy child at term, 26 weeks after the FMT.
Samples were analyzed using Cmbio’s Clonal-level Microbiomics™️ platform. Single nucleotide variants were identified in the metagenomic samples and were compared to a collection of samples from healthy infants and adults. Two forms of engraftment profiling were used, allowing researchers to track microbial populations from FMT donor to recipient—despite the recipient already having closely-related endogenous strains.
Study Results
Results showed that, before receiving FMT, the pregnant mother’s gut microbiota was characterized by low diversity and the presence of potential pathogens. Upon receiving FMT, these features diminished. After the infant’s birth, the precise bacterial strains administered to the mother via FMT were found in the infant, suggesting both persistence over time and cross-generational transfer of the strains.
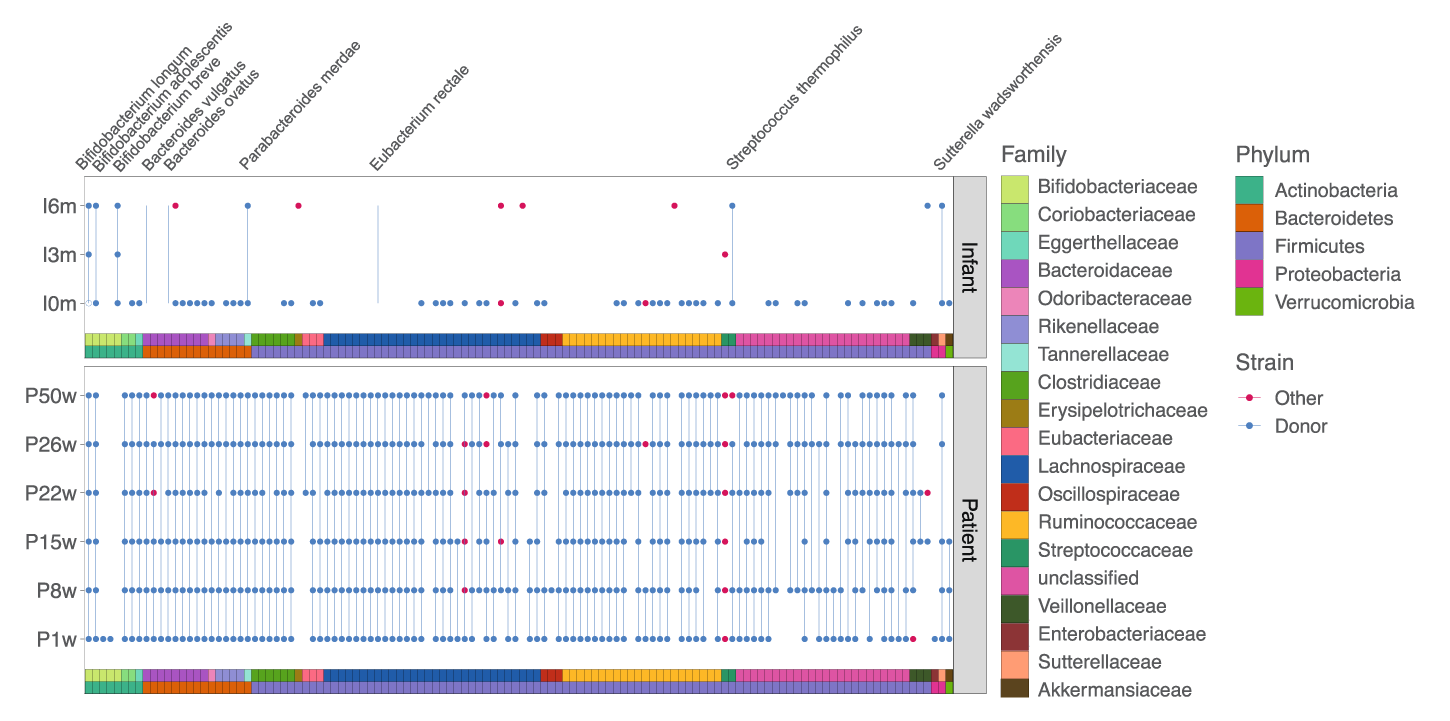
Detection of donor (blue dots) and non-donor (red dots) strains longitudinally in the mother and infant. None of these strains were detected in the mother pre-FMT.
See the study published in *Microbiome.* ¹
This case study demonstrates the potential to track precise strains from one environment to another with confidence, and to determine the persistence of these strains over time. Vertical transfer of bacterial strains from a pregnant mother to an infant was observed, demonstrating a potential route for influencing the infant gut microbiota.
Let’s find the answers. Let our expert scientists guide you. Contact us to discuss your study design and research needs on engraftment and strain tracking.
Reference
¹ Wei, S., Jespersen, M.L., Baunwall, S.M.D. et al. Cross-generational bacterial strain transfer to an infant after fecal microbiota transplantation to a pregnant patient: a case report. Microbiome 10, 193 (2022). https://doi.org/10.1186/s40168-022-01394-w
